Running R from the command line
tl;dr
Run your jobs from bash Rscript --save my_script_file.r instead of doing a source('my_script_file.r') from the R shell.
The problem
I’ve been keeping a casual eye on PCIC’s systems lately, watching the way that people use our computational resources. I’ve been working directly with users a lot less over the last couple of years, so I do more and more monitoring using system tools.
I have noticed lately that a lot of people have a propensity for executing long-running, memory intensive jobs on R’s interactive shell and then leaving them lying around idle before they come back and attend to the results. This is a bad practice for a number of reasons.
Let’s look at the output of the top command below. Notice that userAAA is consuming a huge amount of RAM (look at the “RES”) column. If you add it up, it’s on the order of 40GB which is, like, a third of the size of the disk drives that we have in our desktop machines. But if you look at the CPU column? The processes are not active, not actually doing anything, and just hanging out, waiting for someone to come back to their desk.
PID USER PR NI VIRT RES SHR S %CPU %MEM TIME+ COMMAND
26368 userAAA 20 0 11.9g 11g 6496 S 0 9.2 5:07.11 R
26347 userAAA 20 0 10.1g 9.8g 6428 S 0 7.8 4:15.58 R
26382 userAAA 20 0 4886m 4.4g 6656 S 0 3.5 5:54.27 R
26340 userAAA 20 0 4886m 4.4g 6656 S 0 3.5 5:59.54 R
20631 userBBBB 20 0 3745m 3.6g 3556 S 0 2.8 826:24.54 R
26337 userAAA 20 0 3398m 3.0g 8216 S 0 2.4 4:55.16 R
25027 userBBBB 20 0 2893m 2.7g 3912 R 100 2.2 27:14.06 R
26354 userAAA 20 0 2261m 1.9g 6656 S 0 1.5 5:53.53 R
26389 userAAA 20 0 2261m 1.9g 6656 S 0 1.5 5:47.01 R
26375 userAAA 20 0 2147m 1.8g 6660 S 0 1.4 6:02.08 R
26361 userAAA 20 0 2126m 1.8g 6652 S 0 1.4 5:39.64 R
25660 userCCC 20 0 996m 859m 5160 S 0 0.7 1:28.31 R
Why would someone do this?
I can see the motivation. Climate data is really big, and depending on what you’re doing, it can take a lot of wall time to run through processing all of your data or performing some computation. Sometimes it takes hours. Sometimes it takes days. And this is a research process; one doesn’t necessarily know what will be found in the results, and one doesn’t necessarily know whether the computation was written correctly (“was there a bug in my code?”). So I get it. You need to check on the results afterwards and do some examination.
And don’t get me wrong… we want people to use our system resources. Because climate data is so big, we purchase a tonne of RAM for our systems. This enables users to run big jobs, put as much stuff in RAM as possible, and speed up processing by factors of thousands. So, obviously we want people to use RAM. That’s just being smart, and enabling faster iterations on your research questions.
Using vs. wasting
The problem, however, is that this modus operandi is not using RAM, it’s wasting RAM. atlas and medusa are shared by about 20 users, and when someone is locking the RAM unnecessarily, no one else has it available to do their computation faster. What I have been seeing is that people are running large RAM jobs in the interactive shell that take hours to complete. If something takes hours to complete, you’re not going to attend to it. So the computation completes, and the shell just sits there idle, doing nothing, holding all of your intermediary variables and intermediary data in memory.
For example, right now process 4102 is reserving 2 GB of RAM (along with about about 8 siblings that are each reserving their own set of 2 GB). You may think that it’s doing something important. But if we look at the output of strace:
strace -p 4102
Process 4102 attached - interrupt to quit
select(7, [6], [], NULL, {2590146, 359294}
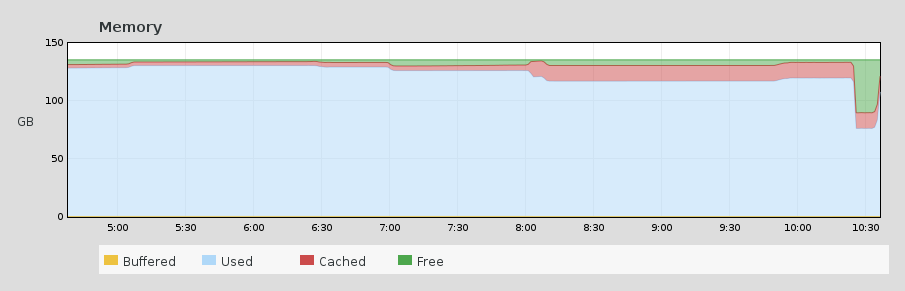
it’s just sitting idle there waiting for input or for some event to happen. That is what the select() system call does. Looking at the timeseries of RAM usage on atlas.pcic, I was able to see exactly when this user came back from our staff meeting and found his/her overnight job finished (and closed down his/her session). Look for the big drop in usage.
A brief aside
Some of you might be asking: if I don’t use the RAM, won’t it just go to waste?
In short, no. The Linux Operating System automatically utilizes all unused RAM for buffer cache. Buffer cache is potentially used to cache any file I/O operations, so that re-reading data from disk can be faster and so that writing data to disk doesn’t have to block the process. Basically when there’s more free RAM, there’s a higher likelihood of your file I/O being cached in RAM by the OS, thus making everything run faster. It’s much complicated than that, but that’s the gist of it.
The solution
So how to we use RAM during our job run and then release it when we’re done? Use Rscript. I’ve seen a lot of people write R script files, open a command line session and then start by typing:
> source('my_script_file.r'). From the perspective of code reuse, this is certainly way better than just typing things into the shell. You can actually go back and see what you ran, make modifications and run it again. But if your script is long running or resource intensive, you are much better off to run it as a batch process with Rscript. When you do, the process gets closed when it finishes and the resources get released. If you need to inspect the results interactively afterwards, or play around on the R shell, just use the --save option, and your whole environment will get saved to disk. Then you can load it back again when you’re ready to attend to your work.
For example, let’s say that I have a script file that adds some numbers together, and suppose that hypothetically it takes a long time.
james@basalt ~ $ cat my_script_file.r
x <- seq(10)
y <- seq(11, 20)
z <- x + y
james@basalt ~ $
I can just run this on the bash command line with Rscript, and tell it to save my environment afterwards.
james@basalt ~ $ Rscript --save my_script_file.r
james@basalt ~ $
Notice that it just runs the job quietly, and produces no output on standard out. It just saves the state to disk and closes down. By default Rscript saves the session to a file named “.RData” in the local directory, but this is also configurable. When I’m rested up, have had a shower and am ready to get back to my work, I can just fire up R and if it finds an .RData file it will automatically load up the previously saved environment.
james@basalt ~ $ R
R version 3.2.0 (2015-04-16) -- "Full of Ingredients"
Copyright (C) 2015 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
Natural language support but running in an English locale
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
[Previously saved workspace restored]
>
If we take a look at what’s in the environment, you’ll see everything that was in our script file.
> ls()
[1] "x" "y" "z"
> x
[1] 1 2 3 4 5 6 7 8 9 10
> y
[1] 11 12 13 14 15 16 17 18 19 20
> z
[1] 12 14 16 18 20 22 24 26 28 30
>
You do need to be careful using this workflow, and only use it when you honestly have to inspect your results. You don’t necessarily want to go dropping 40GB files all over the place, or you’re going to run yourself out of disk space too.
Conclusions
PCIC has a lot of system resources at our disposal for an organization of our size. Thank goodness for that. But please try and be cognizant of how you are consuming those resource. Please don’t be wasteful, because doing so can impact the research of other system users.
When running long R jobs that are resource intensive, use Rscript --save and ensure that your processes get closed down when processing is finished.
Thanks to Basil Veerman for reviewing an ealier draft of this post.
blog comments powered by Disqus